Evaluating the Binding Interactions between Artemisinin and Kelch 13 Protein Mutants Via Molecular Modelling and Docking Studies
DOI:
https://doi.org/10.37934/araset.28.3.272286Keywords:
Malaria, artemisinin, Plasmodium falciparum parasitesAbstract
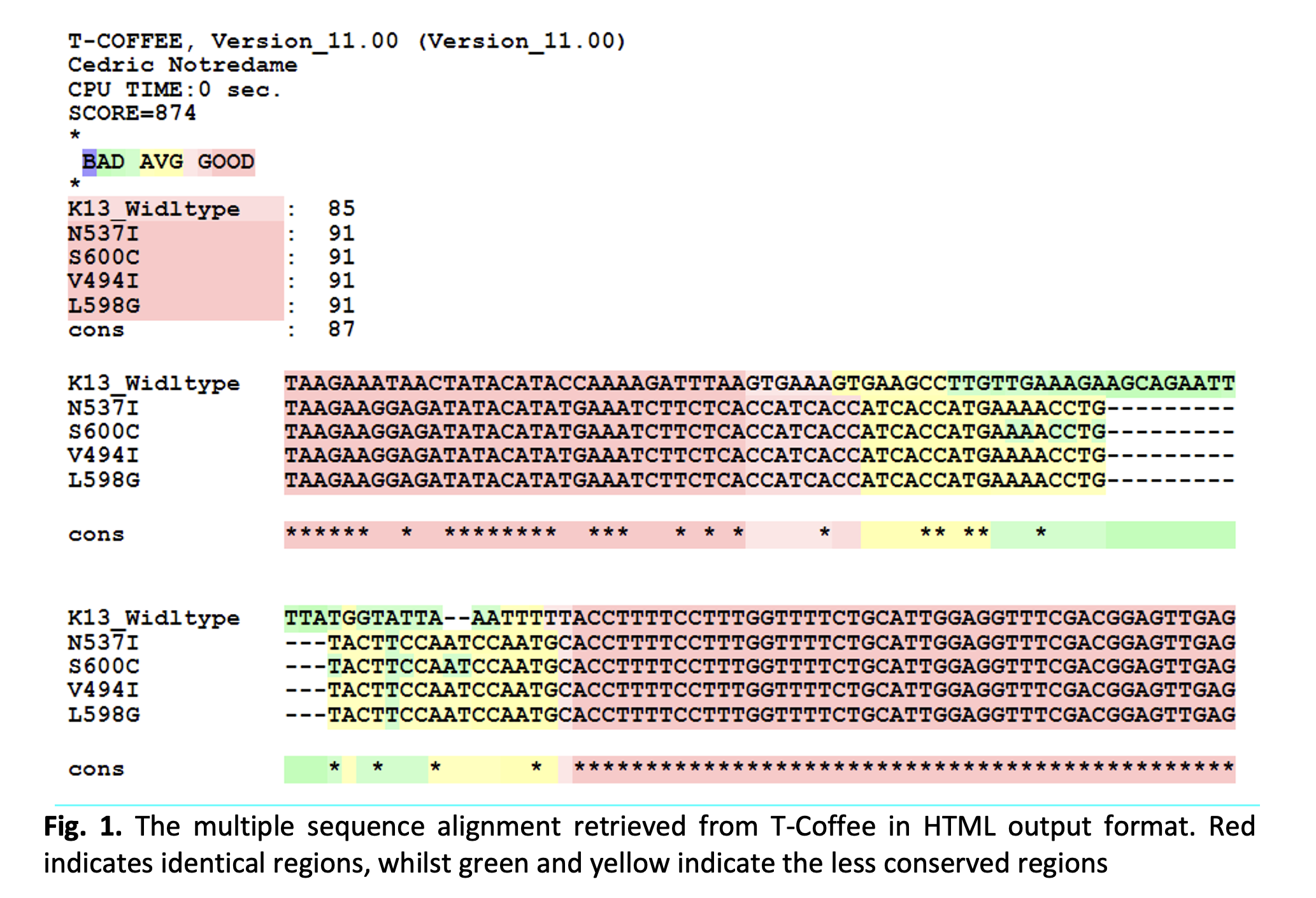
Malaria is a parasitic infection caused by protozoan parasites from the genus Plasmodium. Over the years, various concerns have arisen regarding the efficacy in treating malaria caused by Plasmodium falciparum, which was reported to be caused by mutations in one of the parasite’s proteins, known as the Kelch 13 (K13). This study aims to generate the model structures of P.falciparum K13 protein mutants and to evaluate the binding affinities and interactions between these proteins and artemisinin drug, which is the drug used for the treatment of malaria. To date, the interactions between the protein mutants and artemisinin drug have not been computationally elucidated. In this study, four different types of mutant proteins were analysed, which are V494I, L598G, S600C and N537I and the results were compared with the wild-type K13 protein. Homology models of these proteins were created using the wild-type K13 (PDB ID:4YY8), with high percentage of sequence identity with the mutants. Most models with -2 and 2 have good Rama-Z scores, hence it can be deduced that the four mutants V494I (-1.21 ± 0.42), L598G (-1.19 ± 0.41), S600C (-0.93 ± 0.43), N537I (-1.16 ± 0.43) and the wild-type (-1.34 ± 0.45) have acceptable Rama-Z scores. Molecular docking between artemisinin and the generated models of K13 proteins revealed that all protein mutants have higher binding energy; V494I (-6.79 kcal/mol), L598G (-9.26 kcal/mol), S600C (-6.17 kcal/mol) and N537I (-6.96 kcal/mol), compared to the wild-type (-9.65 kcal/mol). The results showed that all four distinct mutant proteins have less stable complex formation, which indicate that the mutant proteins have higher resistance towards artemisinin due to the higher binding energy compared to the K13 wild-type protein. However, all mutations have a higher number of protein-ligand hydrophobic interactions and protein-ligand hydrogen bonds than the wild-type protein, which requires further analysis to understand the binding interactions. The predicted structural information with regards to binding interactions between the K13 mutant proteins and artemisinin obtained from this study has paved the path toward understanding how mutants may cause parasites’ resistance towards artemisinin drugs.Downloads
Downloads
Published
2022-11-30
How to Cite
Aisya Nazura Azman, Safwan Fathi Muhd Fahmirauf, Fazia Adyani Ahmad Fuad, Raden Shamilah Radin Hisam, & Noor Azian Md. Yusuf. (2022). Evaluating the Binding Interactions between Artemisinin and Kelch 13 Protein Mutants Via Molecular Modelling and Docking Studies. Journal of Advanced Research in Applied Sciences and Engineering Technology, 28(3), 272–286. https://doi.org/10.37934/araset.28.3.272286
Issue
Section
Articles